






Overview
The program-controlled quantitative sealing machine is a specialized device designed to meet modern packaging production needs. It is driven by intelligent circuits and has a dedicated heating roller inside. It is specifically used for sealing 51 well and 97 well quantitative plates, and is used in conjunction with microbial detection reagents to provide a simple, fast, and accurate experimental plan for quantitative detection of total coliforms, fecal (heat-resistant) coliforms, and Escherichia coli; The 51 well quantitative disk and 97 well quantitative disk are semi-automatic quantitative methods designed based on the traditional Maximum Possible Number (MPN) statistical model, which can quantitatively detect water sample results. This method can replace and outperform traditional (membrane filtration, and multi tube fermentation) methods for detecting microorganisms in water.
Scope of application:
Used for rapid detection of total coliforms, fecal (heat-resistant) coliforms, and Escherichia coli in water samples, portable in the field, emergency, and quantitative testing.
Execution standards:
Compliant with GB 3838-2002 Surface Water Environmental Quality Standards
Compliant with CJ/T 206-2005 Urban Water Supply Quality Standards
Compliant with GB/T 5750.12-2023 Standard Test Method for Drinking Water
Complies with the "CJ/T51-2018 Urban Sewage Quality Standard Inspection Method - Determination of Heat Resistant Coliforms - Enzyme Substrate Method"
Complies with the "HJ 1001-2018 National Environmental Protection Standard for Determination of Total Coliforms, Fecal Coliforms, and Escherichia coli in Water Quality"
Product Description:
The enzyme substrate detection device consists of four parts: a programmable quantitative sealing machine, a 51 or 97 well quantitative detection disk, a 100mL quantitative bottle, and an enzyme substrate detection reagent. The detection water sample range includes drinking water, source water, bottled water, reclaimed water, secondary water supply, pipeline water, wastewater, food water, livestock water, medical water, etc.
Advantages and Characteristics:
The enzyme substrate method is a standard method for detecting Escherichia coli included in GB5750-2006. It is currently the most advanced method for detecting Escherichia coli in water, and is gradually being recognized by domestic testing departments for its convenience, speed, low false positives, and suitability for rapid detection of large amounts of samples.
Compared with multi tube fermentation and membrane filtration methods, enzyme substrate method greatly reduces the detection steps and does not require high experimental environment. The detection time can be reduced to 24 hours, which has good application prospects in daily water sample monitoring and emergency monitoring. It can detect and prevent in a timely manner, minimizing the probability of major public safety accidents.
Method advantages:
1. No need to operate in a sterile room.
2. The manual operation time is less than 1 minute.
3. No need for culture medium preparation and extensive sterilization of glassware.
Qualitative and quantitative analysis can be completed in 4.24 hours without the need for validation experiments.
MMO-MUG culture medium using fixed substrate technology and enzyme substrate method in accordance with GB5750-2006 "Standard Test Methods for Drinking Water" is paired with a programmable quantitative sealing machine, and the water sample is packaged with (51 well quantitative disk) or (97 well quantitative disk) for detection.
[Working principle] Add a water sample containing total coliform bacteria, and cultivate the target bacteria in Minimum Medium ONPG-MUG medium at 36 ℃± 1 ℃. The specific biological enzyme β - galactosidase produced by total coliform bacteria can decompose the chromogenic substrate ONPG in Minimum Medium ONPG-MUG medium, making the culture appear yellow; At the same time, Escherichia coli in the water sample produces specific β - glucuronidase to decompose the fluorescent substrate MUG in the Minimum Medium ONPG-MUG medium, producing characteristic fluorescence. Similarly, the heat-resistant coliform bacteria (fecal coliform bacteria) will decompose the chromogenic substrate ONPG in the Minimum Medium ONPG-MUG medium when cultured at 44.5 ℃, resulting in a yellow color of the medium.
Product Features
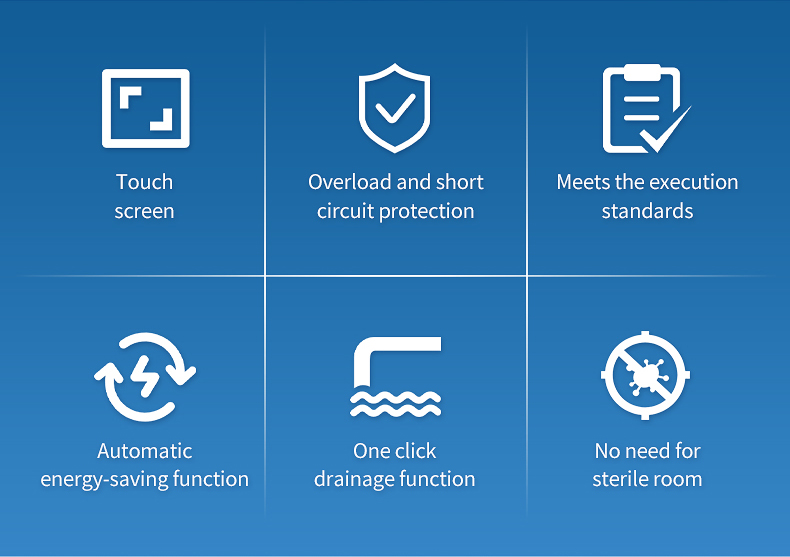
Efficient and energy-saving: Optimized the layout of heating elements and temperature control algorithms, which not only improves work efficiency but also reduces energy consumption.
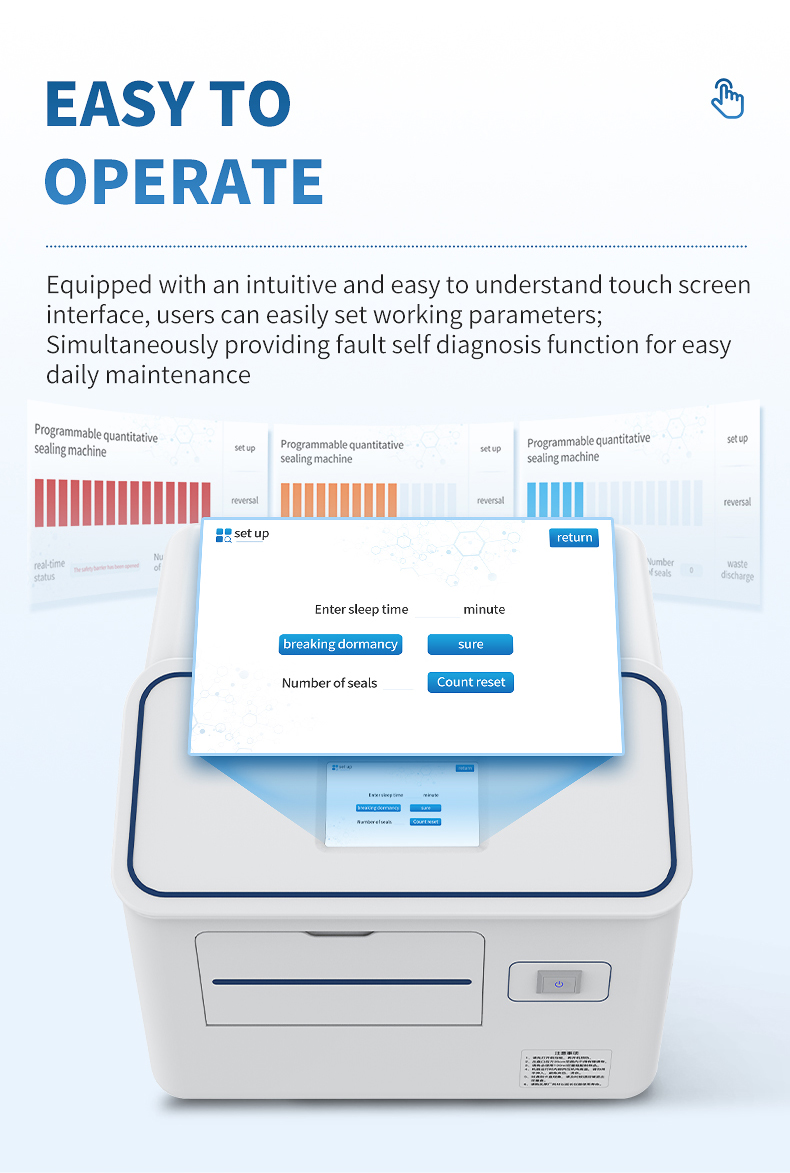
Easy to operate: equipped with an intuitive and easy to understand touch screen interface, users can easily set working parameters; Simultaneously providing fault self diagnosis function for easy daily maintenance.
Safe and reliable: equipped with multiple safety measures such as overload protection and short circuit protection.
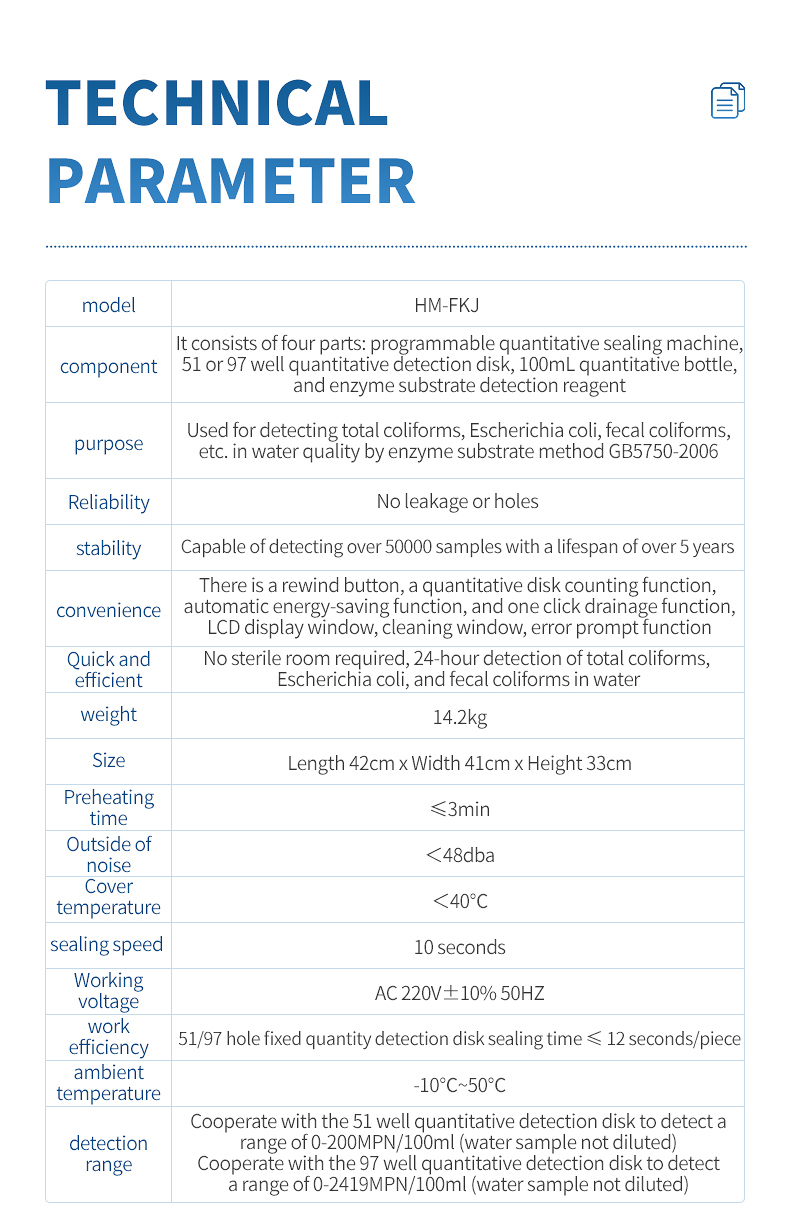
Technical parameters
Usage: Used for GB5750-2006 enzyme substrate method to detect total coliforms, Escherichia coli, fecal coliforms, etc. in water quality
Reliability: No leakage, no holes
Stability: capable of detecting over 50000 samples, with a service life of over 5 years
Convenience: It has a rewind button, a quantitative disk counting function, automatic energy-saving function, one click drainage function, a large LCD display window, a cleaning window, and error prompt function.
Convenience: No sterile room required, 24-hour detection of total coliforms, Escherichia coli, and fecal coliforms in water
Weight: 14.2kg
Dimensions: 42cm in length, 41cm in width, and 33cm in height
Preheating time: ≤ 3 minutes
Noise: < 48dba
Outer shell temperature: < 40 ℃
Sealing speed: 10 seconds
Working voltage: AC 220V ± 10% 50HZ
Work efficiency: Sealing time for 51 hole and 97 hole fixed quantity detection disks ≤ 12 seconds/piece
Working environment temperature: -10 ℃~50 ℃
Detection range: 0-200MPN/100ml (undiluted water sample) when used with a 51 well quantitative detection disk, 0-2419MPN/100ml (undiluted water sample) when used with a 97 well quantitative detection disk
Article address:https://www.molecularbio.cn/pro5/20.html